Work package 1 (WP1): Crop performance
Lead France (FR) with Belgium (BE) and Sweden (SE). Responsibilities: Management of field research units (FRU), data acquisition and analyses will be done by each partner.
Objectives: Evaluate (1) agronomic performance, resources allocation and use efficiency of perennial grain crops in comparison with annual crops, (2) incidence of management practices (including N fertilization, hay-cut, pasture) on above-ground biomass, dual production of forage and grain and ecosystem services, (3) sustainability potential of IWG Intermediate Wheatgrass (IWG; Kernza) according to existing strategies designed with farmers and stakeholders.
Tasks (T): Organise the access to the central field research units (FRU) and deliver relevant soil, plant and climatic data to all WP using established long-term field experiments. This includes the preparation and distribution of soil and plant samples obtained at each FRU.
(T1) Throughout the experimental phase, soil and climatic data will be monitored (e.g. determination of soil texture, primary pools and availability of soil C, N and P, Hot Water Extractable C (HWEC), Particulate Organic Matter). Crop monitoring will consider above-ground biomass, grain and forage yield, and quality determination. Weeds and fungal diseases will be monitored. Root traits and development will be measured over time on FRU3. Particular attention will be paid to the preparation and distribution of soil and plant samples obtained at each FRU.
(T2) The incidence of crop management will be measured on crop performance and ecosystem services. It includes evaluation of N fertilization (FRU1, FRU2) legumes association (FRU, FRU2) and interaction of treatments (Fertilisation*hay-cut on FRU2 and FRU3). N content analyses will be performed on plant biomass samples to understand the N balance in relation to soil N pools and N fertilization regime.
(T3) A system perspective based on life cycle assessment (LCA) will be developed to account for the environmental impacts of different matter fluxes occurring during the process of producing IWG compared with annual crops (e.g. greenhouse gas GHG emissions, potential N and P leaching).
Synergies between tasks: WP1 provides soil samples for WP 2-4, and essential inputs for crop modelling (WP5). WP1 will be the primary platform for co-creation process with farmers and stakeholders (WP6).
Work package 2 (WP2): Crop-associated microbiome
Lead: Poland (PL) with Austria (AT) Responsibilities: Greenhouse experiments in PL and AT, data acquisition and analyses will be done by each partner. Partner SE will conduct complementary analyses with the WP.
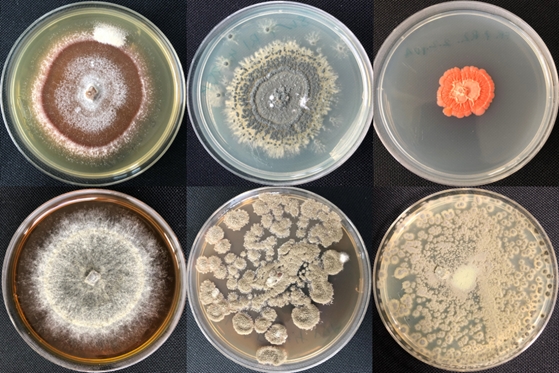
Objectives: (1) Assess bacterial and fungal microbiomes and the genetic signatures of microorganisms associated with IWG grain cropping systems of the NAPERDIV FRU and contrast this with microbial functioning found in combination with annual grain crops; (2) analyse the efficacy of selected bacterial and fungal biostimulants (with K-strategic characteristics) from perennial systems to improve germination and early plant growth (seedling stage) under controlled and field conditions; (3) characterise potential biocontrol agents associated with perennial grain crops. We expect a more stable microbiome in general as well as a higher abundance and diversity of beneficial microorganisms (bacteria, fungi) in perennial systems compared to such found with annual grain crops and with this, a higher ecosystem resilience.
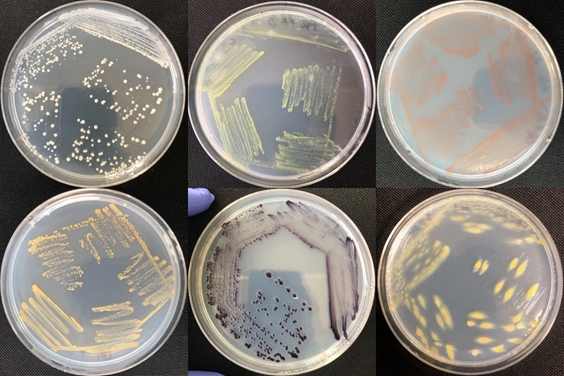
Tasks (T): Samples from crop fields (plant material, rhizosphere and reference soil) cultivated under the tested perennial agricultural conditions will be collected, analysed and tested.
(T1) Samples will be subjected to high-throughput sequencing to determine the taxonomic composition (amplicon sequencing of marker genes) and functioning (shotgun-based sequencing of total community DNA). Bioinformatics will resolve the community structures (QIIME2, R-statistics) and functional repertoires (DIAMOND BLAST against NCBInr, KEGG assignments) focusing on genes associated with biogeochemical element cycling and pathogenesis. Standardisation of sample preparation, data processing as well as long-term sample and data storage will follow the recently developed and validated procedures provided by the EC-funded “Microbiome Support Action”. This will guarantee that generated datasets will be accessible for research groups beyond NAPERDIV. Complementary network analysis will be used to determine community connectivity and interactions, particularly those with inoculants and soil fauna (WP4).
(T2) The impact of grain crop growth and yield parameters will be evaluated upon application of biostimulants (focus on K-strategic microorganisms) that were isolated from perennial systems and subsequently enriched. The selection process will be complemented by using existing strains from a collection of plant-beneficial microorganisms in AT and PL.
(T3). Based on the results from the greenhouse trials, the most promising bacterial and fungal candidates will be tested in the FRU under field management conditions (WP1). Proportions and temporal dynamics of the applied inoculants strains as related to the overall microbiome in the rhizosphere and endosphere (including the seed endosphere as a recently identified hotspot for plant-beneficial microorganisms) of host plants will be assessed during the entire IWG life cycle. In addition, samples will be analysed in terms of bacterial activity and community characterisation by phospholipid fatty acid analysis (PFLA) (WP3).
Synergies between tasks: WP2 will provide relevant contributions to the design of perennial grain cropping systems including a “cropping system prototype” to be developed by WP1. Outputs will serve as a base for novel bio-based solutions to increase microbial biodiversity and associated beneficial traits in crop production.
Work package 3 (WP3): In situ climate change simulation
Lead: Sweden (SE) with Austria (AT) and Poland (PL). Responsibilities: SE for WP coordination, FRU1 (Sweden) management, in situ rain exclusion installation, sampling, data acquisition and soil analyses.
Objectives: WP3 will investigate plant and microbial resistance to recurring drought events against the current scenario of climate change in a perennial grain crop compared with an annual grain crop. Objectives of WP3 include: (1) installation of an in situ rain exclusion equipment in sub-plots within the SAFE facility (FRU1); (2) assessment of provisioning services (above- and below-ground biomass, grain yield and quality, forage yield and quality, harvest index), including analyses of soil physico-chemical properties (pH, organic matter, total C, N and P content, moisture, temperature); (3) analysis of the natural abundance of 13C in shoot and grain tissue to estimate the discrimination against 13C as indicator of water use efficiency; and (4) analysis of the bacterial activity via the leucine incorporation method, soil microbial community structure, biomass of bacteria, saprotrophic and mycorrhizal fungi using phospholipid fatty acid (PLFA) analysis in the rain exclusion sub-plots in contrast to those without rain exclusion. Subsamples of the simulation experiments will also be subjected to DNA-based microbiome profiling (WP2).
Tasks (T): To investigate the resistance of the cropping systems and the soil microbiome against climate change field simulations of drought recurring drought periods.
(T1) Installation of rain exclusion as 2 × 2 m subplots in the main perennial and annual grain crop plots to simulate drought events during summer under two consecutive years. Soil sensors will monitor the moisture and temperature in subplots with and without rain exclusion shelters.
(T2) Assessment of perennial and annual cereal crop performance in terms of grain and aboveground biomass under different water resource availabilities. It is expected that the perennial system will show a stronger drought resistance than the annual systems, due to their inherently extensive root system.
(T3) Analysis of the soil microbiome emphasizing soil microbial community structure, biomass and activity. After one and two growing seasons under simulated drought, both PLFA and NLFA based soil microbial community structures and biomass will be determined, and leucine incorporation will be included as a microbial activity parameter. It is expected that the soil microbial biomass is more resilient (K-strategic behavior) against drought than its activity (r-strategic feature).
Synergies between tasks: Soil sample analyses of WP3 will provide extended knowledge of drought effects on the soil microbiome (WP2) and for its trophic interactions (WP4). Soil samples from WP2 will be shared with WP3 for PLFA/NLFA and microbial activity analyses. Results on crop production of the drought experiment will be prepared for and transferred to WP5 to set up the cropping system model, together with outputs from WP1.
Work package 4 (WP4): Soil fauna diversity
Lead: Germany (DE) with Romania (RO). Responsibilities: University of Hohenheim (DE) for soil-dwelling fauna and overall data analysis and integration, University of Trier (DE) for soil-borne fauna identification and analysis, Sapientia Hungarian University of Transylvania (RO) for soil dwelling fauna identification.
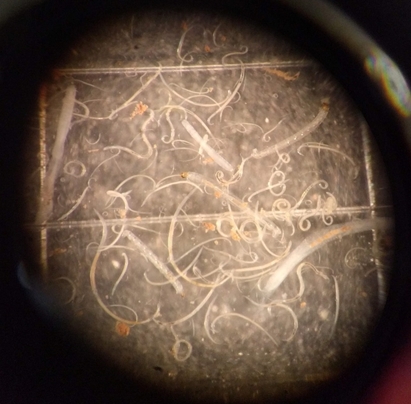
Objectives: (1) Analyse species diversity (species composition and abundance) and functional diversity (food-web relationships) of soil-borne engineers and decomposers (nematodes, earthworms), under IWG in comparison to adjacent annual crop plots; (2) analyse trophic interactions between soil microorganisms and the free-living, non-parasitic nematode assemblage to estimate potential impacts on bacterial and fungal microbiomes (assessed in WP2) and their related soil quality parameters (assessed in WP1); (3) analyse species diversity and abundance of ground- and rove beetles to estimate the potential of pest suppression in IWG compared to annual systems; and (4)develop and adapt ecological indicator schemes for nematodes and the selected ground-dwelling beetle taxa to estimate their potential for assessments of biodiversity (including species conservation value) and yield potentials in perennial cropping systems.#
Tasks (T): For representative nematode, earthworm and beetle species recording, we make use of three study regions of contrasting climatic conditions in Europe, i.e. SE (FRU1), BE (FRU2) and FR (FRU3), which provide IWG plots of appropriate size and age (BE sites are not available for beetles).
(T1) Soil-borne nematode fauna will be investigated from representative mixed soil samples and subsequent Baermann funnel method. Nematodes will be identified on a family/genus level. Nematode sampling will be conducted once per study region and treatment. Changes in the nematode assemblage will be calculated using the Maturity Index (M.I.), the Enrichment (E.I.)- and the Structure (S.I.)-Index gaining nematode faunal profiles.
(T2) Earthworms will be extracted from soil in spring and autumn each year according to DIN EN ISO 23611-1. We will estimate earthworm abundance, biomass, species diversity, and life-form strategy, and thus, species-related functional traits (as far as possible). These indicators will allow us to evaluate changes in life history, feeding characteristics and community stability of perennial grain cropping systems.
(T3) Ground- and rove beetles beetle (Carabidae and Staphylinidae) recording will be done by using pitfall traps (Barber traps) in a standardised approved pitfall trap design. Based on the ecological behaviour of the selected taxa (peak occurrence), recording will be conducted over 8 weeks each in spring and additionally after crop harvest (autumn) in two consecutive years. Species will be identified, additional catches of other arthropds such as spiders will also beconsidered. To finally identify factor-species correlations among beetle communities, multivariate statistics will be used.
(T4) Selected taxa will be utilised as ecological indicators to assess the overall biodiversity status and ecosystem services of perennial grain cropping systems. We will use a novel framework based on the identification of indicator species for single or multiple ecosystem services across taxonomic groups, using a recently developed indicator species analysis approach deploying multi-species community data.
Synergies between tasks: WP4 will provide essential knowledge on the functional biotic structure and interactions of the soil fauna related to determined soil properties, soil-based ecosystem services and crop performance attributes (WP1). This will be complemented with the study of specific interactions between nematodes and the crop-associated microbiome (WP2) as well as selected soil parameters, such as pH, SOM, nutrients, and bulk density. These data will be integrated in the general framework with practical guidelines to direct further co-creation process of innovative nature-based cropping systems (WP6) and be used for development of an indicator system for soil fauna diversity and related ecosystem services such as natural pest control.